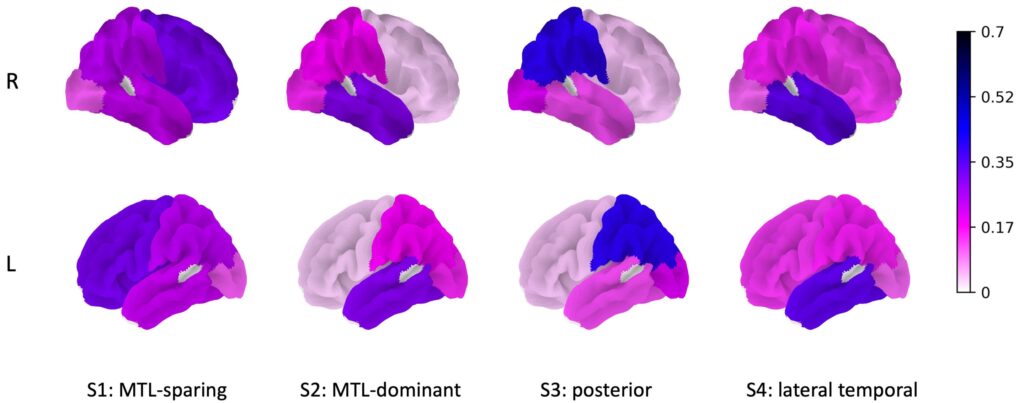
A new diagnostic method that combines genomic and tau PET imaging promises a way to personalize Alzheimer’s disease. Based on the combined knowledge structure using sparse canonical correlation analysis (SCCA), the combined method was able to identify four subtypes of Alzheimer’s disease and the top genes associated with them.
The research was presented at the 2023 Society of Nuclear Medicine and Molecular Imaging Annual Meeting and was published in Journal of Nuclear Medicine.
Alzheimer’s disease is a genetic disorder. Several subtypes of the disease have been identified based on PET imaging of amyloid plaques and tau neurofibrillary tangles, hallmarks of the disease. In addition, recent genome-wide association studies have identified many genes that contribute to the development of Alzheimer’s disease.
“By identifying multiple forms of Alzheimer’s disease using imaging and genomic information, researchers can gain new insights into the causes and progression of the disease,” said Joyita Dutta, Ph.D., assistant professor in the Department of Biomedical Engineering. at the University of Massachusetts in Amherst, Massachusetts. “Understanding the specific interactions of each species may also lead to future interventions.”
Imaging and genomics from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) were used in the study. All ADNI participants met 18F-flortaucipir PET and Illumina SNP genotyping were combined (541 participants in total; 334 cognitively normal and 207 cognitively impaired). Standardized tau PET based on the number of trees from 10 broad regions was calculated from tau PET imaging, and 145 genome-wide variants associated with Alzheimer’s disease were identified and excluded from SNP genotyping. The SCCA-clustering procedure was applied to both tau PET and genomics datasets.
Four areas of Alzheimer’s disease were identified in the analysis: medial temporal lobe (MTL)-dominant, posterior, MTL-sparing, and lateral-temporal. In addition to the APOE gene, top genes associated with each subtype were also identified.
“Genomics- and imaging-guided individualization is important in Alzheimer’s disease because different subtypes may also have different rates and histories of cognitive decline, which may affect the results of clinical trials and response to treatment,” said Dutta. “By combining molecular knowledge and genomics, we have developed a diagnostic method that can be personalized for each patient. This can be useful in the diagnosis of many types of diseases, not just Alzheimer’s disease.”
More information:
Abstract 1377. “An SCCA-clustering framework for Alzheimer’s disease subtyping using tau PET and genomics,” Fan Yang and Joyita Dutta, University of Massachusetts, Amherst, Massachusetts; Matthew Maher and Richa Saxena, Massachusetts General Hospital, Boston, Massachusetts.
More news:
Journal of Nuclear Medicine
Presented by the Society of Nuclear Medicine and Molecular Imaging
#Genomics #imageguided #subtyping #guide #characterization #Alzheimers #disease